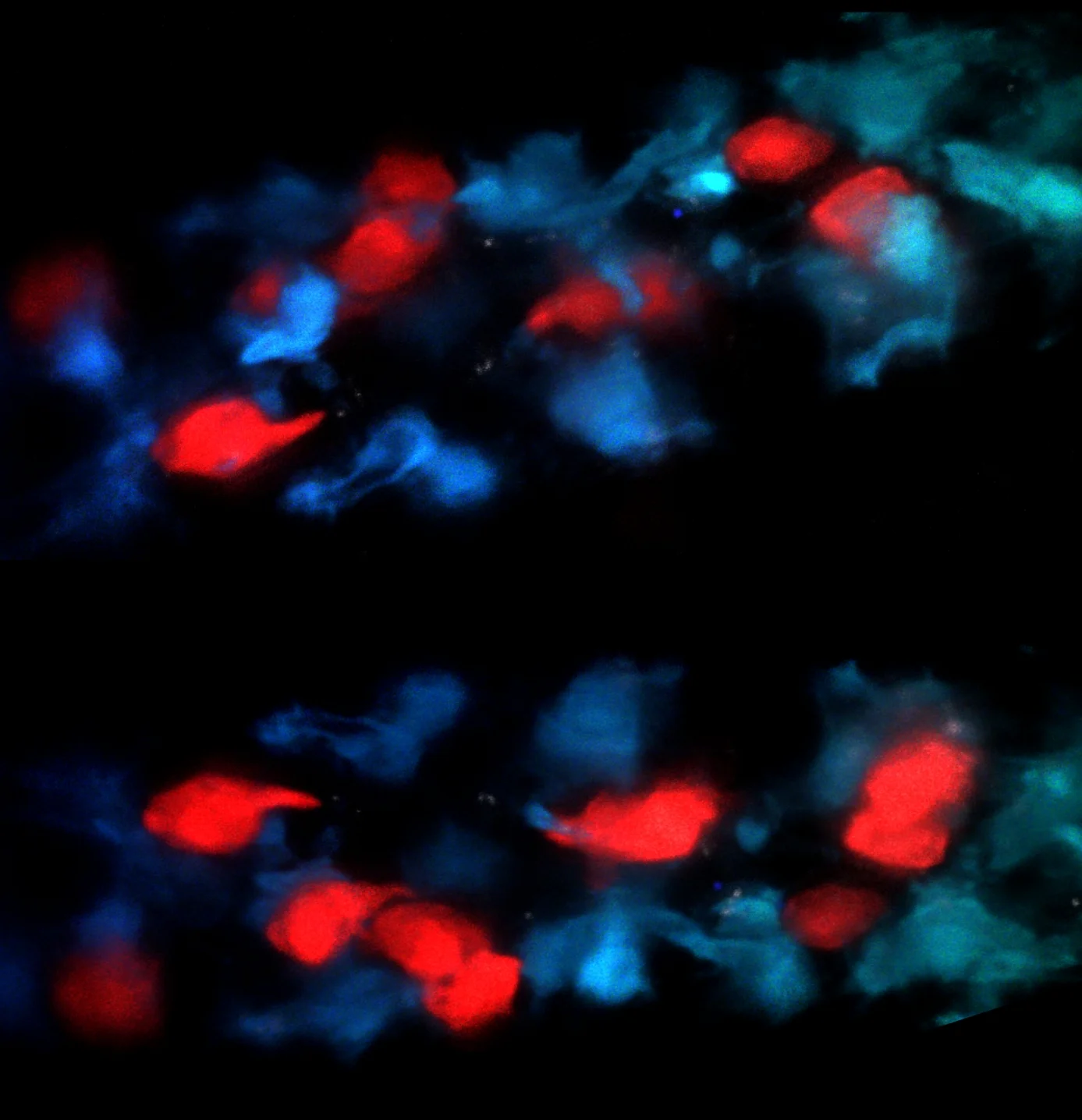
Microbial Regulation of Mucosal Healing
One part of our lab focuses on understanding the cellular and molecular mechanisms of homeostatic inhibition/induction by microbes within the intestine. One type of mononuclear phagocyte (MNP) in the intestine which expresses the fractalkine receptor CX3CR1 plays a critical role in this process. We, and others, have shown that in the steady state, commensal microbiota support the tolerogenic function of these cells within the intestine. However, under conditions of stress and altered microbiota that occur in IBD, these CX3CR1+MNPs can traffic luminal microbes to the mesenteric lymph nodes and stimulate aberrant responses to commensals. Under conditions of colitis, these MNPs expand in the lamina propria and, we have recently shown, support a critical function of innate lymphoid cells (called ILC3s) to produce the cytokine IL-22 and promote mucosal healing. Notably, CX3CR1+ MNP highly express TL1A/TNFSF15 whose gene encodes numerous single nucleotide polymorphisms (SNPs) associated with IBD. Acting via the monogamous receptor DR3/TNFRSF25, TL1A potently enhances the production of effector cytokines by binding to its monogamous receptor DR3/TNFRSF25 on ILC3. Critical questions remain regarding the nature of the bacterial signals impacting these cells, the local and molecular signals regulating these cellular interaction, and the in vivo role for DR3-signaling in ILC3-dependent homeostasis. Using patient-derived microbiota as well as mouse models with unique reporters, conditional genetic deletions, or potential for targeted cellular ablation, our labs aims to understand the signals critical to promoting barrier function.
Two to tango. Mononuclear Phagocytes interact with innate lymphoid cells in the intestinal villi.
Microbial Regulation of Joint Inflammation in IBD
A second part of the lab focuses on microbial changes associated with extra-intestinal manifestations (EIMs) of IBD and their influence on mucosal and systemic immunity. In particular, seronegative spondyloarthropathies (SpA) designate a group of diseases with overlapping genetic and clinical features in which the gut is the putative port of entry for microbial triggers of systemic cellular inflammation resulting in joint disease. Thus, studying patients with IBD-associated SpA gives us a unique opportunity to understand the process by which luminal microbes shape systemic immune repertoires. Consistent with the hypothesis that luminal microbial triggers can impact inflammatory disease, we recently performed a cross-sectional analysis of fecal microbiota composition in patients with new onset rheumatoid arthritis (NORA) compared to chronic RA, psoriatic RA, and healthy controls. Interestingly, the results showed an expansion of P. copri in patients with NORA and pangenome analysis by shotgun sequencing identified potential marker genes associated with disease phenotype. Interestingly, the inverse correlation of P. copri abundance with the RA genetic susceptibility allele HLA-DRB1 suggests that P. copri itself may confer inflammatory response or serve as a diagnostic surrogate for an inflammatory risk factor. We hypothesize that key luminal microbiota exist in the intestine of patients with IBD-associated SpA and contribute to disease. Drawing off our large clinical IBD practice at the Jill Roberts Center for IBD, we are using innovative clinical phenotyping and 16S rRNA as well as metagenomic sequencing to define microbial signatures associated with disease. Anaerobic culturing has enabled us to isolate arrays of unique, patient-derived microbial clones, which are used to probe systemic immunityin vitro as well as their effects on immune cell function in gnotobiotic mice.
bum Joints. Intestinal Bacteria link mucosal and systemic immunity.